Xylose Lysine Deoxycholate (XLD) Agar is a selective medium for the isolation of Salmonella and Shigella spp from clinical specimens and food samples. XLD Agar was originally formulated by Taylor for the isolation and identification of Shigella from stool specimens.
The pathogens are differentiated not only from the non-pathogenic lactose fermenters but also from many non-pathogens which do not ferment lactose or sucrose.
Additionally, the medium was formulated to increase the frequency of growth of the more fastidious pathogens, which in other formulations have often failed to grow due to the inclusion of excessively toxic inhibitors.
The results obtained in a number of clinical evaluations have supported the claim for the relatively high efficiency of XLD Agar in the primary isolation of Shigella and Salmonella. XLD Agar is included in the USP microbial limit test for screening specimens for the presence or absence of Salmonella and is recommended for the testing of foods, dairy products and water.
Principle of XLD Agar
XLD Agar is both a selective and differential medium. It contains yeast extract as a source of nutrients and vitamins. It utilizes sodium deoxycholate as the selective agent and, therefore, is inhibitory to gram-positive micro-organisms. Xylose is incorporated into the medium since it is fermented by practically all enterics except for the Shigella and this property enables the differentiation of Shigella species. Lysine is included to enable the Salmonella group to be differentiated from the non pathogens since without lysine, salmonellae rapidly would ferment the xylose and be indistinguishable from non-pathogenic species. After the salmonellae exhaust the supply of xylose, the lysine is attacked via the enzyme lysine decarboxylase, with reversion to an alkaline pH which mimics the Shigella reaction.
To prevent similar reversion by lysine positive coliforms, lactose and sucrose are added to produce acid in excess. Degradation of xylose, lactose and sucrose to acid causes phenol red indicator to change its colour to yellow. Bacteria that decarboxylate lysine to cadaverine can be recognized by the appearance of a red colouration around the colonies due to an increase in pH. These reactions can proceed simultaneously or successively, and this may cause the pH indicator to exhibit various shades of colour or it may change its colour from yellow to red on prolonged incubation.
To add to the differentiating ability of the formulation, an H2S indicator system, consisting of sodium thiosulfate and ferric ammonium citrate, is included for the visualization of the hydrogen sulfide produced, resulting in the formation of colonies with black centers. The non pathogenic H2S producers do not decarboxylate lysine; therefore, the acid reaction produced by them prevents the blackening of the colonies which takes place only at neutral or alkaline pH.
Uses of XLD Agar
- XLD Agar is a selective differential medium for the isolation of Gram-negative enteric pathogens from fecal specimens and other clinical material.
- It is especially suitable for the isolation of Shigella and Salmonella species.
- Microbiological testing of foods, water and dairy products.
Composition of XLD Agar
Ingredients per liter of deionized water (Hardy Diagnostics XLD Agar)
| Lactose | 7.5 gm |
| Sucrose | 7.5 gm |
| Sodium Thiosulfate | 6.8 gm |
| L-Lysine | 5.0 gm |
| Sodium Chloride | 5.0 gm |
| Xylose | 3.75 gm |
| Yeast Extract | 3.0 gm |
| Sodium Deoxycholate | 2.5 gm |
| Ferric Ammonium Citrate | 0.8 gm |
| Phenol Red | 0.08 gm |
| Agar | 15.0 gm |
Final pH 7.4 +/- 0.2 at 25 degrees C.
Preparation of XLD Agar
- Suspend 55 grams of dehydrated medium in 1000 ml purified or distilled water.
- Heat with frequent agitation until the medium boils.
Note: DO NOT AUTOCLAVE. - Transfer immediately to a water bath at 50°C.
- After cooling, pour into sterile Petri plates.
Note: It is advisable not to prepare large volumes, which will require prolonged heating and may produce precipitate.
Colony Characteristics of XLD Agar
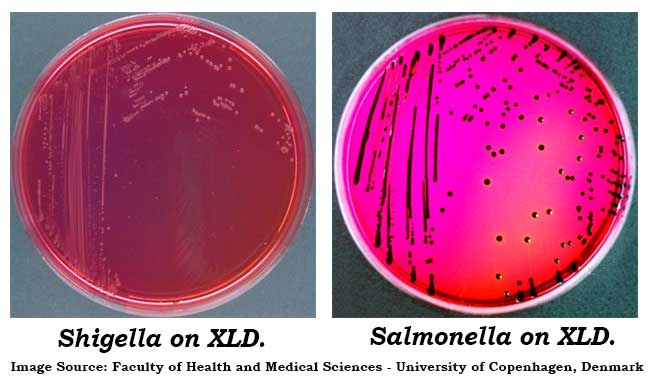
- Degradation of xylose, lactose and sucrose generates acid products, causing a color change in the medium from red to yellow.
- Hydrogen sulfide production under alkaline conditions causes colonies to develop black centers. This reaction is inhibited by the acid conditions that accompany carbohydrate fermentation.
- Lysine decarboxylation in the absence of lactose and sucrose fermentation causes reversion to an alkaline condition and the color of the medium changes back to red.
Typical colonial morphology on XLD Agar are as follows:
Salmonella Typhi – Red Colonies, Black Centers
Salmonella choleraesuis – Red Colonies
Shigella sonnei – Red Colonies
Shigella flexneri – Red Colonies
Escherichia coli – Large, Flat, Yellow Colonies; some strains may be inhibited
Proteus vulgaris – Yellow Colonies
Enterobacter/ Klebsiella – Mucoid, Yellow Colonies
Pseudomonas aeruginosa – Pink, Flat, Rough Colonies
Gram-positive bacteria – No growth to slight growth
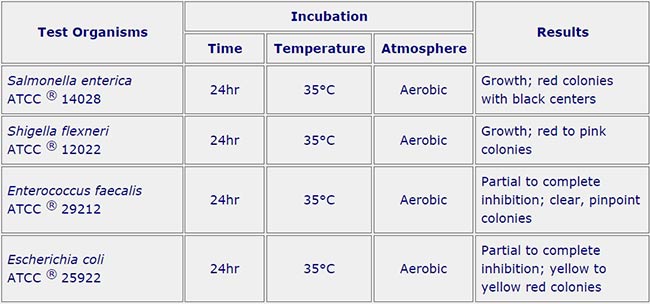
Quality Control for XLD Agar
 Limitations of XLD Agar
Limitations of XLD Agar
- Red, false-positive colonies may occur with some Proteus and Pseudomonas species.
- Incubation in excess of 48 hours may lead to false-positive results.
- S. paratyphi A, S. choleraesuis, S. pullorum and S. gallinarum may form red colonies without black centers, thus resembling Shigella species.
- Some Proteus strains will give black-centered colonies on XLD Agar.
- For identification, organisms must be in pure culture. Morphological, biochemical, and/or serological tests should be performed for final identification. Consult appropriate texts for detailed information and recommended procedures.
- A single medium is rarely adequate for detecting all organisms of potential significance in a specimen. Cultures of specimens grown on selective media should, therefore, be compared with specimens cultured on nonselective media to obtain additional information and help ensure recovery of potential pathogens.




ليست هناك تعليقات:
إرسال تعليق
Due to the high number of spammy comments we have decided to initiate comment moderation so that we can maintain our quality standards and make good environment for our visitors. Please leave your comment